Linda Kenney, PhD
Professor, Department of Biochemistry & Molecular Biology
Professor, Professor of Microbiology and Immunology
Tom and Kaye Arnold Professor in Gastroenterology
Tel: (409) 747-0387
E-mail: likenney@utmb.edu
Campus Location: 6.136 Medical Research Bldg
Mail Route: 0645
Research
Salmonella can adopt either a virulent or a dormant lifestyle inside its host. What makes Salmonella such an expert pathogen is its ability to switch lifestyles depending on the environmental conditions that it encounters. Under favorable
conditions (e.g. the acid pH of the vacuole), Salmonella activates its repertoire of virulence genes, which allows it to invade host cells and actively spread infection (Liew et al., eLife 2019). Under neutral conditions, dormancy genes are
activated that allow the bacteria to lay low and form clusters called biofilms. The two-component response regulator SsrB acts canonically through phosphorylation from its kinase SsrA to activate virulence genes, and non-canonically when the SsrA
kinase is at very low levels to de-repress H-NS and drive activation of the biofilm master regulator CsgD (Desai et al. eLife, 2016), reviewed in (Desai & Kenney, 2017, 2019). Dormant states have been characterized by the formation of biofilms
inside the host and the molecular mechanisms that lead to the formation of biofilm inside live worm hosts were identified in our recent study (Desai et al., PNAS 2019) (see Figure). This work uncovers how the molecular mechanisms that drive biofilm
formation under laboratory conditions are equally relevant in living systems and it now allows us to translate laboratory knowledge into tools for tackling bacterial persistence and pathogenesis in living hosts. 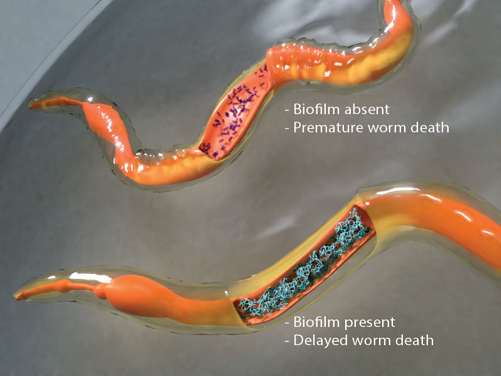
Our most recent project involves developing the oncolytic potential of Salmonella. It is capable of targeting tumors and causing tumor regression. In order to understand the fundamentals of tumor targeting and regression, we are combining spheroids, organoids and heterologous host models, including: C. elegans, Zebrafish and the mouse Lastly, our laboratory has a long-standing interest in signal transduction and the regulation of gene expression in prokaryotes. In particular, we study the two-component regulatory system EnvZ/OmpR that regulates the expression of outer membrane proteins, as well as many other genes involved in acid and osmotic stress. Our present work focuses on how OmpR activates genes required for systemic infection located on Salmonella pathogenicity island 2 in Salmonella enterica.